hindmarsh-rose-2d
Hindmarsh-Rose lattice model in Python and julia
To cite this code, please use:
This repo contains implementations of the Hindmarsh-Rose model of neuronal excitability on a 2D lattice.
The code is available in Python and Julia.
Ref.: Hindmarsh, J.L., Rose, R.M., A model of neuronal bursting using three coupled first order differental equations. Proc R Soc Lond B 221:87-102, 1984.
Rendered page: https://frederic-vw.github.io/hindmarsh-rose-2d/
Python Requirements:
- python installation, latest version tested 3.6.9
- python packages (
pip install package-name)- NumPy
- Matplotlib
- opencv-python (save data as movie)
Julia Requirements:
- julia installation, latest version tested 1.6.1
- julia packages (
julia > using Pkg; Pkg.add("Package Name"))- NPZ
- PyCall (load Matplotlib animation functionality)
- PyPlot
- Statistics
- VideoIO (save data as movie)
Hindmarsh-Rose model
The Hindmarsh-Rose model uses three variables to model membrane potential dynamics in response to current injections. The model is an extension of the FitzHugh-Nagumo model. The third variable of the Hindmarsh-Rose model implements slow inactivation dynamics of the membrane potential and allows to model neuronal bursting. Spatial coupling is introduced via diffusion of the voltage variable ($D \Delta V$):
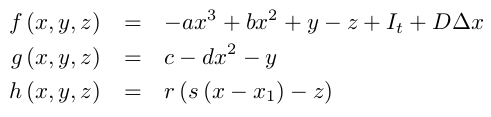
Noise is added via stochastic integration of the variable $x$:
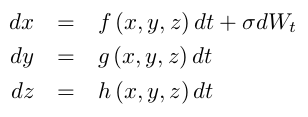
The main function call running the simulation is: hr2d(N, T, t0, dt, sd, D, a, b, c, d, s, r, x_1, I0, stim, blocks):
N: lattice size(N,N)T: number of simulation time stepst0: number of ‘warm-up’ iterationsdt: integration time stepsd: noise intensity (σ)D: diffusion constanta,b,c,d,s,r,x_1: Hindmarsh-Rose model parameters,I0: stimulation current amplitudestim: stimulation current parameters, array of time-, x-, and y-interval bordersblocks: conduction blocks, array of x- and y-interval borders
Outputs: (T,N,N) array as NumPy .npy format and as .mp4 movie.
In example 1, use
stim = [ [[0,15000], [5,N-5], [5,N-5]] ]blocks = [](works in Python, in Julia change 0 to 1)
Example-1
Stimulation of a large block of the simulation area generates a depolarization in that area in which different wave patterns develop, including spiral waves.
Parameters:
N = 64, T = 15000, t0 = 2500, dt = 0.05, sd = 0.05, D = 2.0, a = 1.0, b = 3.0, c = 1.0, d = 5.0, s = 4.0, r = 0.001, x_1 = -1.6, I0 = 3.5
Conclusions
The Hindmarsh-Rose lattice model can produce multiple wave patterns.